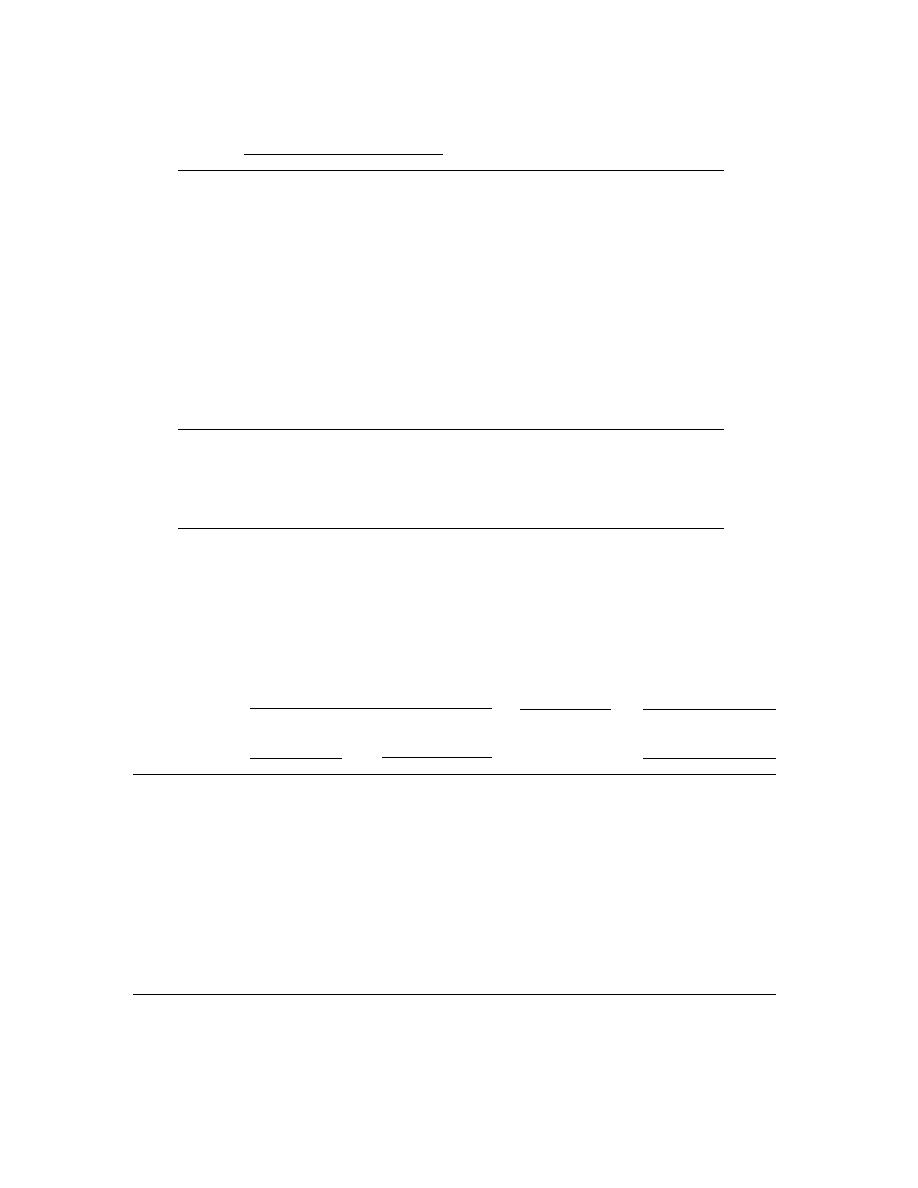
Table 4. Comparison of HMX concentration estimates (mg/kg) using the on-site
colorimetric results for individual subgrids, mean of four subgrids, and grid com-
posites.
Subgrid
Subgrid
Grid no.
A
B
C
D
Mean
Composite
% Diff.*
variability**
D1
120
121
32
59
83
80
3.6
3.8
D2
279
271
316
41
227
212
6.6
7.7
D4
631
148
254
680
428
479
11.9
4.6
D5
53
33
86
54
57
41
28.1
2.6
D7
209
1000
729
1140
770
812
5.5
5.5
D8
878
286
202
208
394
322
18.3
4.3
D9
10
180
20
14
56
35
37.5
18.0
D10
25
5.1
2.4
6.4
9.8
4.7
52.0
10.4
C4
1910
721
995
2160
1447
1636
13.1
3.0
C5
461
79
129
301
242
180
25.6
5.8
Means
371
380
Mean (all)
6.6
Mean (except D9 and D10)
4.7
t = 0.382†
Paired t-test (Composite vs. subgrid mean)
% Diff. Mean (all values)
20.2%
% Diff. Mean (conc. > 50)
14.1%
* Absolute value of [100% (composite/subgrid mean 100)].
** Highest subgrid concentration/lowest subgrid concentration.
† Composite and mean of subgrids not significantly different at 99.9% confidence level.
Table 5. Comparison of measures of analytical precision, accuracy and discrete sample representa-
tiveness for CFB-Valcartier (CFB-V) results relative to those found for other explosives analytes at
Monite (M), Hawthorne (H), and Volunteer (V).
Precision
Accuracy
Local heterogeneity
Largest
Ratio of highest mean
concentration
Slope of
concentration vs. lowest
Sample
RSD of duplicates
ratio of duplicates
0-intercept model
for discrete samples
location
On-site
Lab
On-site
Lab
On-site vs. lab
On-site
Lab
CFB-V (HMX)
6.9
19.2
1.818
2.537
0.988
20.2
34.3
CFB-V (TNT)
**
**
1.6
3.6
1.051*
>7
>7
M-1 (TNT)
3.9
11.1
1.157
1.473
0.815
243
315
M-2 (DNT)
23.0
10.0
1.655
1.461
0.350
10.6
33.4
M-3 (TNT)
16.7
6.5
1.822
1.186
1.464
50.0
98.1
H-4 (TNT)
12.5
13.5
1.696
1.986
0.911
69.0
58.1
H-5 (TNT)
3.3
4.9
1.126
1.157
0.847
28.9
29.5
H-6 (picrate)
11.6
11.9
1.500
1.875
0.967
688
43,000
V-7R (TNT)
4.9
5.1
1.265
1.214
0.677
3.8
3.0
V-8 (TNT)
19.7
4.5
1.731
1.185
1.070
53.1
55.6
V-9 (TNT)
4.1
5.1
1.131
1.167
1.032
8.2
5.7
Mean
(TNT only)
9.3
7.2
1.418
1.338
0.983
65.1
80.7
* Slope for model with intercept. Intercept was 0.63, but was found to be statistically significant at the 95% confidence
level.
** RSDs could not be computed because many values were less than detection limits for one of the duplicates.
15




 Previous Page
Previous Page
